HSP70: Figures
Figure 1. Domain structure of HSP70s.
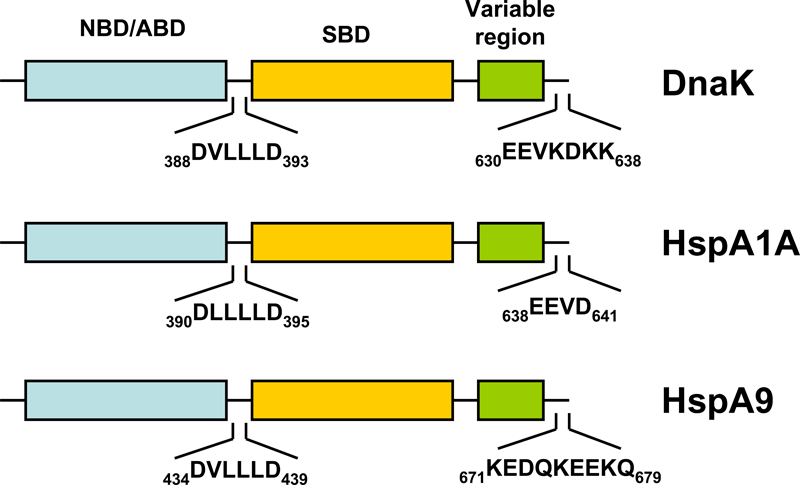
Figure 1. Schematic representation of the domain structure of pro- and eukaryotic HSP70s.
HSP70s are composed of an N-terminal nucleotide binding domain (NBD; also termed ATP-binding domain, ABD) and a substrate binding domain (SBD) followed by a variable region and the C-terminal EEVD-motif. In contrast to the other cytosolic HSP70s, HspA9 (Hsp70-9) lacks the C-terminal EEVD-motif involved in binding of co-chaperones and other HSPs.
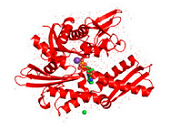
Figure 2. Conformations of human Hsp70-1.
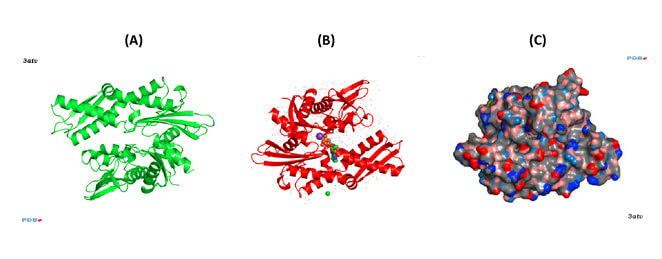
Figure 2. Conformations of human Hsp70-1 (PDB ID: 3atv; 299).
Ribbon and tube representation of the tertiary Hsp70-1 structure in the absence (A) and presence (B) of ADP. Non-polymeric entities are as follows: potassium (grey), chloride (green). (C) Electrostatic surface distribution of Hsp70 complexed with ADP. Blue and red colours of the electrostatic surface diagram correspond to positive and negative electrostatic potentials of standard residues in proteins and polynucleotides. Similarly, electro-positive and electro-negative polar atoms are skyblue and salmon, respectively. Default element colours are: carbon (grey), nitrogen (blue), oxygen (light pink), sulphur (green), phosphorus (olive-green), selenium (magenta).
Figure 3. Comparison between the open and closed conformation of Hsp70.
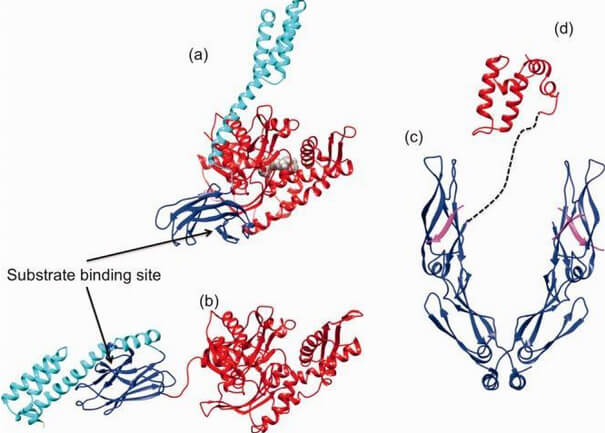
Figure 3. Comparison between the open and closed conformation of Hsp70.
Cartoon and surface representation of the ATP-open conformation (a) of E. coli Hsp70 DnaK (PDB ID: 4B9Q; 54) and the ADP-bound closed (b) conformation (PDB ID: 2KHO; 55). NBD is given in red, SBD in blue, and the C-terminal lid in cyan. (c) Ribbon and tube representation of the tertiary structure of the C-terminal homodimeric Hsp40 peptide-binding fragment (blue) complexed with the Hsp70 C-terminal EEVD peptide (magenta, PDB ID: 3AGY; 60). (d) Structure of the J-domain of Hsp40 (red; PDB ID: 2O37). The dashed line indicates the connectivity of Hsp40 domains (reprinted from Clare and Saibil 300).
Figure 4. The Hsp70/DnaK chaperone cycle.
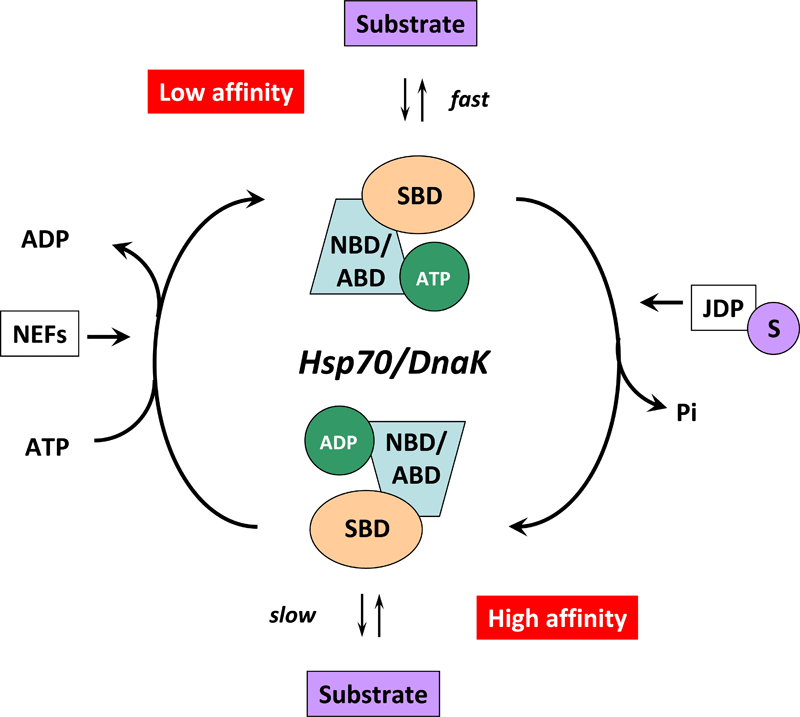
Figure 4. Cartoon illustrating the Hsp70/DnaK chaperone cycle.
In the ATP-bound state, the NBD/ABD and SBD are docked, the substrate binding grove is open, and the Hsp70/DnaK chaperone exhibits low affinity and fast exchange rates for its substrate. ATP hydrolysis is facilitated by J-domains proteins (JDPs) of the HSP40/DNAJ family and nucleotide exchange by nucleotide exchange factors (NEFs). For this purpose, JDPs with the C-terminally bound substrate (S) interact with Hsp70/DnaK. The simultaneous interplay of the substrate with the SBD of Hsp70/DnaK and of the JDP with the NBD/ABD of Hsp70/DnaK induces a conformational change in the ATPase domain thereby stimulating ATP hydrolysis and mediating the closure of the substrate binding grove. In the ADP-bound state, the docking between the two domains of Hsp70/DnaK is interrupted and the chaperone exhibits high affinity and low exchange rates for its substrate. ADP release is mediated by the specific interaction of NEFs with the Hsp70/DnaK ATPase domain followed by a conformational change which results in a low-affinity state and release of the substrate.