HSP70: Structure
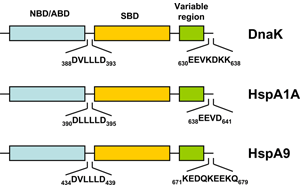
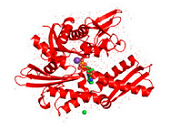
HSP70s are highly conserved and consist of two functional domains (Figure 1 + Figure 2): a 44 kDa N-terminal nucleotide binding domain (NBD; also termed ATP-binding domain, ABD) which binds and hydrolizes ATP and a 28 kDa C-terminal substrate binding domain (SBD) which binds extended polypeptides 48, 49. The ABD is conserved in all of its members with the exception of the two HSPA12 genes, where the ABD is more divergent but still recognizable. The NBD is composed of four subdomains (IA, IB, IIA, IIB) surrounding the ATPase-binding pocket 48, 49.
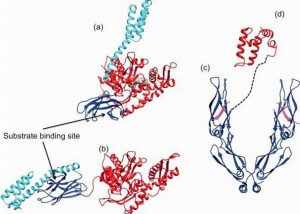
The SBD is subdivided into an N-terminal two-layered twisted β-sheet (SBDβ) and a C-terminal α-helical subdomain (SBDα) providing a flexible lid of SBDβ 50. Eukaryotic cytosolic HSP70s also contain a G/P-rich C-terminal region harboring an EEVD-motif involved in binding of co-chaperones and other HSPs 51. The EEVD-motif is absent from specialized HSP70s such as the ribosome-associated Ssb1/2p in yeast. Members residing in certain subcellular compartments bear an N-terminal localization signal while the ER-specific Hsp70 -5 (HspA5) contains a C-terminal ER retention signal. 52Central to the chaperone function of HSP70s is the transition between open and closed conformations of their SBD (Figure 3).
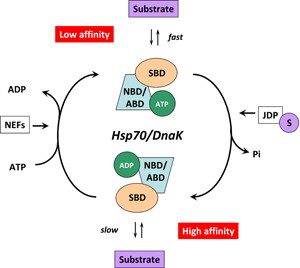
In the ATP-bound open conformation, the β-sheet and α-helical lid subdomains of the SBD are detached from one another and docked to different faces of the NBD giving a low affinity for the substrate 53, 54. Docking of the β-sheet and α-helical lid subdomains to the NBD is a sequential process affected by peptide and protein substrates. In the ADP/peptide state, SBD plus linker collides with and samples the surface areas of subdomains IA and IIA of the NBD and that the IA/IIA cleft is closed 55. ATP binding to the NBD alters the substrate affinity to the SBD and substrate binding to stimulate ATP hydrolysis 53. Substrate-induced ATPase stimulation then leads to SBD release enabling closure of the lid subdomain and fixing of the substrate 55. Cycling between ATP-bound and ADP/substrate-bound states requires HSP70s to visit a state with high ATPase activity and fast on/off kinetics of substrate binding. In the ATP state, the NBD surface groove opens up and allows the NBD-SBD collisions becoming productive, which place the linker in the groove docking the SBD on the IA area 55. Nucleotide exchange factors (NEFs) then participate in ADP/ATP exchange as a prerequisite for re-opening of the lid and release of the substrate 56, 57. HSP70s are assisted not only by NEFs but also by various co-chaperones called J-domain proteins (HSP40s) 58. Members of the HSP40/DNAJ family stimulate the intrinsic ATPase activity of HSP70s and coordinate it with substrate binding by interacting with the Hsp70 ATPase domain through their J-domain (Figure 3) 59. Recruitment and transfer of substrate protein to Hsp70 is not only mediated by interactions with the NBD but also with the SBD of Hsp70. HSP40s also interact with the C-terminal octapeptide of human Hsp70, 634GPTIEEVD641 via a C-terminal peptide-binding domain 60. It is interesting to note that some HSP40s target HSP70 activity to clients at precise locations in cells and others bind client proteins directly, thereby delivering specific clients to HSP70 and directly determining their fate 4. A proposed model of the Hsp70/DnaK chaperone cycle is given in Figure 4.