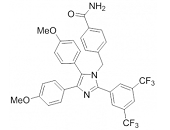
HSP70: Species Variation
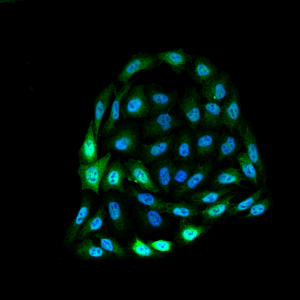
Immunofluorescence detection using mouse anti-bovine HSP70 monoclonal Antibody, clone 1.86 in HeLa cells.
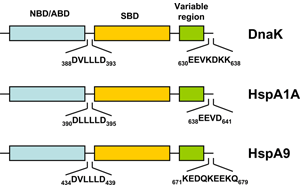
The bacterial homolog of the HSP70 family is referred to as DnaK. E. coli K-12 encodes three HSP70 genes (dnaK, hscA, hscC). Studies during the last years clearly demonstrated that the folding of newly-synthetized cytosolic proteins in E. coli is mainly orchestrated by DnaK in conjunction with the highly conserved molecular co-chaperones trigger factor (TF) and GroEL (Hsp60) 34. Analogous to the other HSP70 family members, DnaK (Figure 1) is a 638 amino acid long protein bearing an N-terminal nucleotide binding domain (NBD) with ATPase activity, a C-terminal substrate binding domain (SBD) subdivided into a β-sandwich subdomain and an α-helical domain forming a lid. Both domains are connected by a highly conserved linker which possesses a characteristic 388DVLLLD393 hydrophobic segment essential for allosteric communication between the NBD and the SBD 35. DnaK also possesses a short conserved region at the C-terminus acting as an auxiliary binding site for unfolded substrates 36. By conferring selective advantage to pathogens, DnaK also plays an essential role in other bacteria including Vibrio cholerae, Streptococcus intermedius, and Staphylococcus aureus thus attesting for its tremendous role in prokaryotic physiology (for a review see 37). A further gene of the HSP70 family of chaperones in E. coli is hscA encoding a 66-kDa protein product (HscA). This gene also encodes a 21-kDa protein with significant homology to the DNAJ protein family 38. HscA is 39% identical and 56% similar to DnaK at the amino acid sequence level 39. HscC, a new DnaK homologue of E. coli encodes a 62-kDa protein sharing 33% homology in primary structure with DnaK and HscA 40. For comparison, the evolutionary relatedness of DnaK to human Hsp70-8 (Hsc70) approaches 50% amino acid identity 39.
A thorough search of the genome sequence of Arabidopsis thaliana revealed 18 members of the HSP70 superfamily in plants, including 14 in the DNAK subfamily (Table 2) 41. The Athsp70 gene products are assigned to distinct subcellular locations. AtHSP70s that contain GPKIEEVD at the C-terminus (AtHsp70-1 to 70-5) are predicted to localize in cytoplasm, whereas those with specific targeting signals at the N-terminus are predicted to localize to specific organelles. While AtHsp70-6 to 70-8 reside in chloroplasts, AtHsp70-9/10 represent the mitochondrium-specific and AtHsp70-11/12 the ER-specific isoforms. Although AtHsp70-13 does not bear an ER retention signal at the C-terminus and the overall sequences show apparent divergence for the last 21 amino acids at the C-terminus, AtHsp70-13 has been assigned to the ER group 41. Most deduced proteins are predicted to locate to the cytoplasm, however, AtHsp70-1, and AtHsp70-5 also contain consensus nuclear localization signals 42. On the other hand, AtHsp70-18 has been found as being most similar to cytoplasmic AtHsp70-1 to AtHsp70-5, but it does not contain the consensus sequence for the cytoplasmic HSP70s in eukaryotes, EEVD, at the C-terminus (Figure 1). Since no expressed sequence tag (EST) was found for this gene and no expression expression could be detected, Hsp70-18 can be considered as a pseudogene 41.
The unicellular green alga Chlamydomonas reinhardtii is somewhat unusual among eukaryotes in that it expresses only a single DnaK-type cytosolic Hsp70 protein along with at least six other HSP70 family members located in different subcellular compartments as well as the flagella 43. In the latter, CrHsp70 obviously plays a crucial role in regulating flagellar assembly and motility 44,45,46. The activity of CrHsp70 also appears to be required in the cytosol for preassembly of dynein arm complexes prior to their transport into the flagellar compartment 47.